Abstract
Livestock productivity can be severely compromised by low fertility due to conceptus loss. Prenatal mortality is of particular concern in swine reproduction. Conceptus loss in pigs occurs mainly in early pregnancy (a primary loss of ~30%) but also during the mid-gestation period (a second loss of ~20%). Autophagy is a degradation system that controls the clearance and reuse of intracellular constituents as well as balancing the sources of energy in times of development and stress. There is compelling evidence to suggest that autophagy plays a role in embryogenesis and pregnancy-allied complications in mammalian species. Hence, the objective of the present experiment was to determine the expression of autophagy-related genes Atg9a and Atg9b in the endometrial and trophoblast tissues of healthy and arresting porcine conceptuses on gestational days 20 (gd20) and 50 (gd50). Relative mRNA expression was assessed by real-time polymerase chain reaction, and protein levels were quantified by Western blot and densitometric analyses. Atg9a mRNA expression was greater (P < 0.05) in the trophoblast from arresting (AT) compared with healthy embryos (HT) on gd20, while Atg9b mRNA expression was greater (P < 0.05) in AT than in HT on gd50. No protein expression was detected with the Western blots of both Atg9 proteins in trophoblast tissue samples on either of the two gestational days except for low levels of Atg9a protein expression in AT samples on gd20; there were no significant differences in Atg9 protein content between tissues and gestational days in the sows of the present study. On the basis of variations in Atg9 mRNA levels in the trophoblast, in can be proposed that the autophagy system is mainly involved in the autoregulation of porcine embryonic/fetal development. Further studies are needed to elucidate the specific roles of intrauterine Atg9 genes and their products throughout porcine pregnancy.
Keywords
Pig; Pregnancy; Conceptus; Endometrium; Trophoblast; Autophagy
Introduction
Prenatal mortality is an important adverse factor that influences reproductive efficiency of livestock [1]. Two waves of spontaneous conceptus loss may occur in porcine pregnancy: a primary embryo loss of ~30% is usually observed around gestational day 20 and a second loss of ~20% of remaining fetuses during mid-gestation (~day 50; [2]). “Crowding” in the uterus due to limited space for developing embryos and fetuses (“uterine capacity”; [2]) or asynchronous development of the uterus and the embryo [1] may both result in the arrest and demise of porcine conceptuses.
Autophagy is a degradation mechanism that controls the clearance or recycling of intracellular constituents as well as balancing the sources of energy during development and stress ([3] Figure 1). The autophagy system is also responsible for removing damaged organelles, clearing non-functional proteins, eliminating pathogens, and recycling misfolded proteins [3]. There is increasing evidence that autophagy plays an important role in mammalian differentiation and development [4]. Autophagy begins with the formation and elongation of phagophores. The phagophores engulf organelles and sequester them in a double-membrane autophagosome [3]. The autophagosome then fuses with the lysosome and matures into autolysosome. The content of the autolysosome is degraded by proteases and the products of degradation are exported into the cytoplasm in order to be reused for various metabolic processes including the biosynthesis of ATP, nucleic acids, carbohydrates, amino acids and lipids. The autophagy processes provide the cell with sufficient nutrients until conditions improve and the cell can survive on its own.
The role of autophagy in pregnancy was studied at the fetal-maternal interface in normal and aberrant human pregnancies [5]. Increased intensity of autophagy processes was observed in the specimens obtained after spontaneous miscarriages; this increase was due mainly to an elevated concentration of autophagy vacuoles. These results can be interpreted to suggest that the conditions inside the uterus of the spontaneous miscarriage enhanced the activity of the autophagy system. Genetic screening has identified a number of autophagy-related genes (Atg genes; [3]). The Atg9 genes and proteins are responsible for the formation of the double-membrane autophagosome; in situations of starvation, the Atg9 vesicles fuse with the outer membrane of the autophagosome [6]. Two isoforms of the Atg9 proteins, Atg9a and Atg9b, are typically required for autophagosome formation [6, 7]. A previous study using the fetal Atg9a knockout mouse model has shown considerable growth restriction and increased mortality of Atg9a-/- fetal mice, suggesting that the non-functional autophagy system can impede or prematurely terminate fetal development [8]. In addition, the intrauterine growth retardation of the Atg9a-/- fetal mice was associated with the presence of maternal hypertension. The aforementioned observations in pregnant women and small laboratory rodents prompted us to examine the pattern of changes in Atg9 gene expression in healthy and arresting porcine conceptuses collected on gestational days 20 and 50.
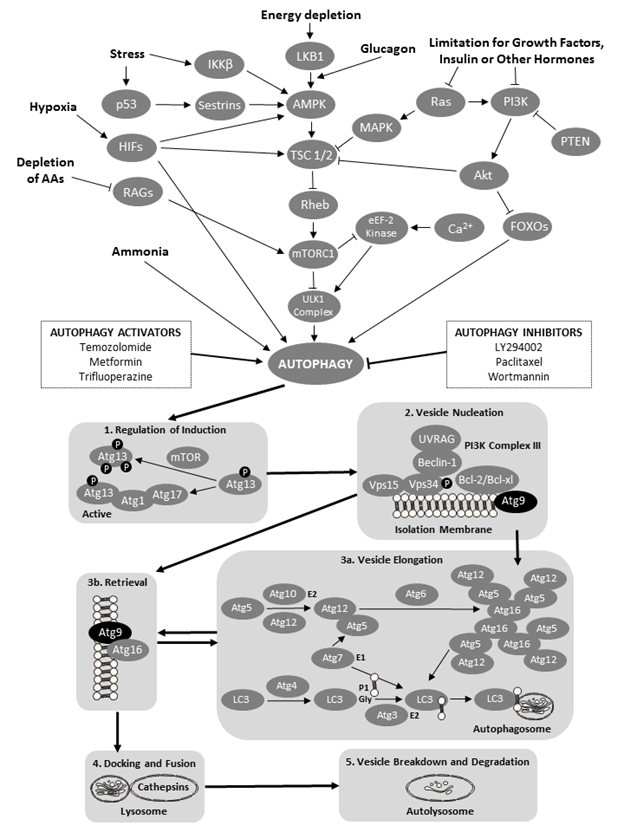
Figure 1. A schematic of the cellular macroautophagy processes (based on a flowchart by the Selleck Co. (Houston, TX, USA) shown here: http://www.selleckchem.com/pharmacological_autophagy.html). Abbreviations used (in alphabetical order and in bold font face for easier identification within the legend): AAs – amino acids; Akt – protein kinase B; AMPK – AMP-activated protein kinase; Atg − autopha-gy-related gene proteins; Bcl – B-cell lymphoma proteins; HIFs – hypoxia-inducible factors; eEF-2 kinase − eukaryotic elongation factor 2 kinase; FOXO − forkhead box protein O; iKK-β − inhibitor of nuclear factor kappa-B kinase subunit beta; LC3 − microtubule-associated protein 1A/1B-light chain 3; LKB1 − liver kinase B1 a.k.a. serine/threonine kinase 11 (STK11) or renal carcinoma antigen NY-REN-19; LY294002 − a morpholine-containing chemical compound that is a potent inhibitor of numerous proteins, and a strong inhibitor of phosphoinositide 3-kinases; MAPK – mitogen-activated protein kinase; mTOR − mechanistic target of rapamycin, formerly mammalian target of rapamycin; mTORC1 − mammalian target of rapamycin complex 1 or mechanistic target of rapamycin complex 1; p53 − tumor protein p53, a.k.a. cellular tumor antigen p53, phosphoprotein p53, tumor suppressor p53, antigen NY-CO-13, or transformation-related protein 53; PI3K − phosphoinositide 3-kinase; PTEN − phosphatase and tensin homolog; RAGs − recombination-activating genes; RAS − small GTPase proteins; RHEB − Ras homolog enriched in brain (GTP-binding protein); ULK1 Complex − serine/threonine-protein kinase complex; UVRAG – UV radiation resistance-associated gene protein; and VPS − vacuolar protein sorting-associated protein. Atg 9 proteins have been denoted by black oval shapes.
Methods and Materials
Second parity Yorkshire sows housed in the Arkell Swine Research Station near Guelph, ON, Canada, were used in this study. All sows were artificially inseminated within 7 days post-weaning (at first estrous detection and 24 h later) using pooled Duroc semen (Ontario Swine Improvement; Innerkip, ON, Canada). Tissue samples were collected from eight pregnant animals on gestational day 20 (gd20) and from eight pregnant sows on gestational day 50 (gd50). The University of Guelph Animal Care Committee had approved animal handling and euthanasia protocols (Animal Utilization Protocol no. 10R061). All experimental procedures were in compliance with the guidelines of the Canadian Council on Animal Care and Use of Experimental Animals. The reproductive tracts were collected immediately after slaughter and within 30 min transported on ice to the laboratory. To remove the embryos from the attachment sites, the uterine horns were opened longitudinally along the anti-mesometrial side. Conceptuses from each sow were categorized as either healthy or arresting based on embryonic/fetal size, weight and visual assessment of vascularization of the attachment sites as previously described [9-11], Figure 2; the conceptuses were removed from the study if they possessed debatable health status or were classified as reabsorbing. Endometrial tissue on the maternal side was collected separately from the trophoblast. All samples were rinsed with PBS and then frozen immediately and stored at −80°C until RNA isolation. The RNeasy mini kits (Qiagen; Mississauga, ON, Canada) were used to perform total RNA extraction from all collected tissues. Total RNA concentrations were determined using the A260/A280 (the ratio of absorbance at 260 and 280 nm) and the Gene Quant pro RNA/DNA calculator (Biochrom Ltd.; Cambridge, UK). Extracted RNA samples were frozen immediately at −80°C. Subsequently, the First-Strand cDNA Synthesis Kit (GE Healthcare Bio-Science Inc.; Baie d’Urfe, QC, Canada) was used for cDNA synthesis. In short, 20 μl of diluted RNA at concentrations ranging from 224 to 890 ng/μl were heated for 10 min at 65°C in the GeneAMP polymerase chain reaction (PCR) System 2700 (Applied Biosystems; Foster, CA, USA). Once the heating was finished, 11 μl of the bulk first-strand cDNA reaction mix, 1 μl of poly (dT) primer and 1 μl of DTT solution was added; the reagents were then incubated at 37°C for 1 h. Using the Gene Quant Pro RNA/DNA calculator, the cDNA concentration was measured. The cDNA products were then stored at −20°C for quantitative real-time PCR.
Primers targeting the genes of interest (porcine Atg9a and Atg9b) were designed using the Primer 3 software from the electronic nucleotide database, GenBank. Aliquots of cDNA were pooled and used as a template to test the primers and to optimize their efficiency. Table 1 lists the primers for β-actin (ACTB), Atg9a and Atg9b. The LightCycler 480 SYBR Green I Master (Roche Diagnostics; Mannheim, Germany) was used to optimize primer efficiency in real-time PCR system (ViiATM 7 Real Time PCR System, Applied Biosystems by Life Technologies; Foster, CA, USA). Genes quantified in the samples were run in duplicates using the MicroAmp Fast 96-well reaction plate (Biosystems by Life Technologies; Foster, CA, USA). Data were analyzed using the ViiATM 7 software for the ViiA 7 Real-Time PCR System (Biosystems by Life Technologies; Foster, CA, USA). All results from the PCR reactions were expressed as a ratio of Atg9a or Atg9b mRNA relative to β-actin (ACTB) mRNA.
For Western blot analyses of Atg9a and Atg9b proteins, the tissues were thawed on ice and divided into 30-mg samples placed in Eppendorf tubes. A protease inhibitor and 200 μl of phosphate buffered saline (PBS) were added to each sample followed by homogenization on ice for 1 min. Homogenates were centrifuged at 4°C for 15 min and the supernatants were separated. The Bradford method was used to determine protein concentrations. The samples were then diluted to a final concentration of 2 μg/μl and stored at −80°C. Protein samples (30 μg) were denatured at 100°C for 5 min using the Applied Biosystems GeneAmp PCR System 2700. Samples were loaded in the wells using the 4-20% Mini-PROTEAN TGX Pre-cast gels (12 well comb/20 μl/well; Bio-Rad Laboratories; Mississauga, ON, Canada). Protein fractions were separated by electrophoresis for 1 h at 140 V. The proteins were transferred onto nitrocellulose membrane by assembling a “gel sandwich” that was immersed in the transfer buffer and exposed to 90 V for 90 min. The membrane was blocked with 2.5 g of skimmed powder milk and 50 ml of Tris-buffered saline with Tween 20 (TBS-T) at room temperature for 1 h and then incubated overnight at 4°C with the primary antibody: Atg9a Goat Polyclonal IgG (sc-70141, Santa Cruz Biotechnologies Inc.; Dallas, TX, USA) or Atg9b Goat Polyclonal IgG (sc-163710, Santa Cruz Biotechnology) diluted 1: 200 in skimmed milk with TBS-T (40 μl of primary antibody with 8 ml of skimmed milk blocking solution). After washing with TBS-T, the membrane was incubated with the secondary antibody: HRP-conjugated rabbit anti-goat IgG (ref-61120; Invitrogen-Thermo Scientific; Frederick, MD, USA) diluted 1: 10,000 in skimmed milk with TBS-T (0.8 μl of secondary antibody with 8 ml of blocking solution) for 1 h at room temperature. Subsequently, the membrane was washed again and imaged using the electroluminescence kit and ChemiDocTM MP Imaging System (Bio-Rad Laboratories; Mississauga, ON, Canada). The membrane was then stripped using the Restore Western Blot Stripping Buffer (Thermo Scientific; Rockford, IL, USA) for re-probing for β-actin; the same protocol as for Atg9a and Atg9b proteins was used except that a primary antibody pre-conjugated with horseradish peroxidase (HPA) was anti-β actin antibody (mAbcam 8226; Abcam PLC; Cambridge, UK). The membranes were imaged and analyzed using the Image Lab analytical software (Version 5.1 Bio-Rad Laboratories Inc.; Hercules, CA, USA) to obtain relative expression values (ratios to β-actin) from each corresponding blot.
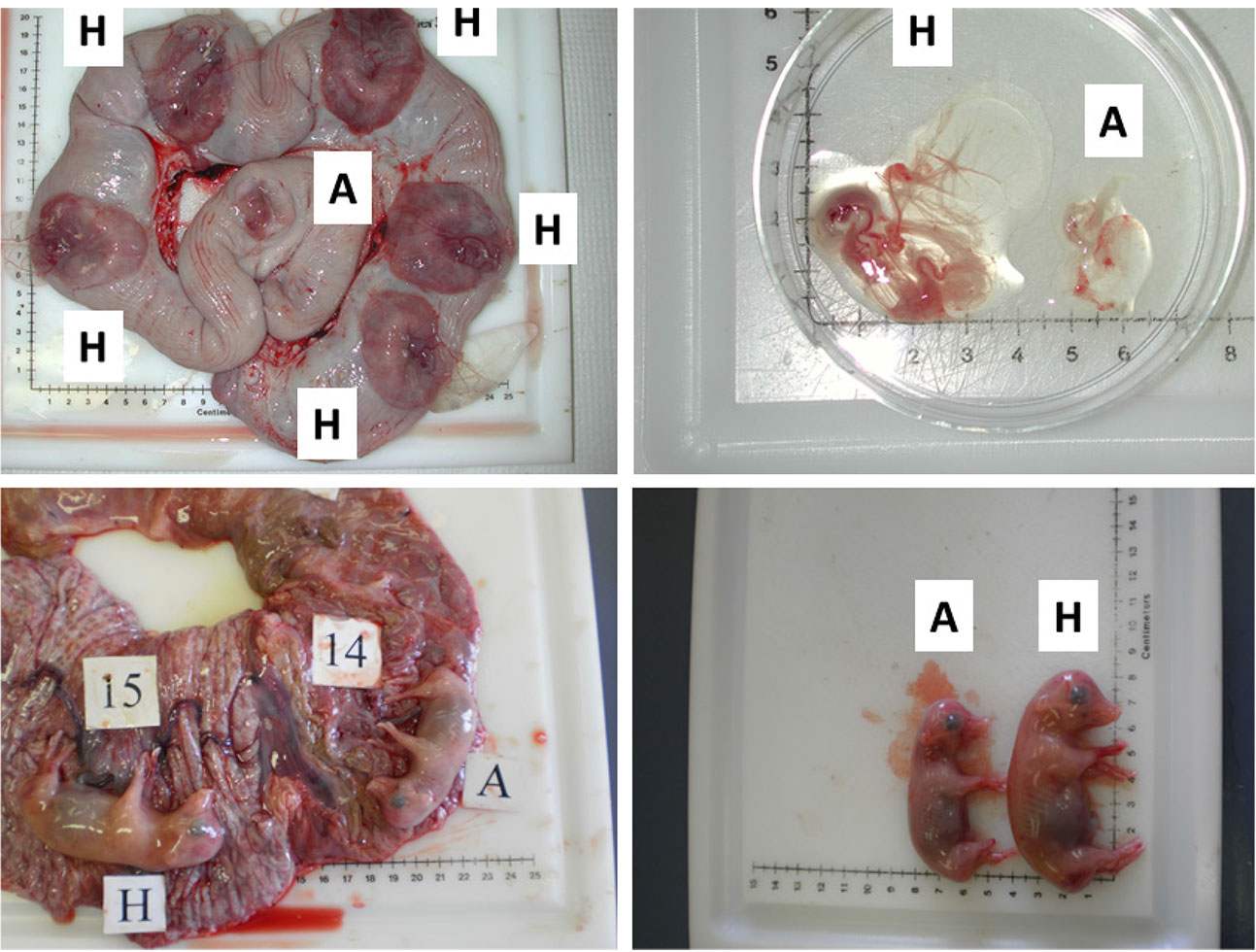
Figure 2. Images illustrating the early (gestational day (gd) 20) and mid-gestational (gestational day (gd) 50) conceptus status in porcine pregnancy. Two upper panels depict healthy (H) and arresting (A) conceptuses on gd20, classified on the basis of disparity in size and vascularization of fetal membranes (Kridli et al., 2016), and two lower panels show healthy (H) and arresting (A) conceptuses from a uterine horn, which were classified on the basis of weight and length on gd50 (Kridli et al., 2016).
Table 1. Specific primers used for Atg9a and Atg9b and β-actin (ACTB) quantification by rtPCR.
| Gene | Primers (5`to 3`) | Product size (bp) |
| Atg9a | Forward: ATCCTCGCTCACATCCACTAC Reverse: GTGAAATTGCGAAGAAGTCTA |
206 |
| Atg9b | Forward: GCATCTGCCGAGATCAGTC Reverse: TCCTTCTGGGTGTCCGTAGT |
266 |
| ACTB | Forward: ACGTGGACATCAGGAAGGAC Reverse: ACATCTGCTGGAAGGTGGAC |
210 |
Statistical analyses were conducted using the SigmaPlot® software (Systat Software Inc., Richmond, CA, USA). To attain normality before statistical testing, all of the data were transformed logarithmically. For real-time PCR and Western blot, the results for endometrial and trophoblast tissue samples across both gestational days were compared by two-way analysis of variance (ANOVA). Comparisons were also made between the tissues obtained from healthy and arresting conceptuses. A P-value < 0.05 was considered significant. All results are given as mean ± standard error of the mean (SEM).
Results and Discussion
Atg9a mRNA expression was significantly greater in arresting than in healthy trophoblast tissue on gestation day 20 (gd20), and it was greater (P < 0.05) in AT on gd20 compared with AT on gestation day 50 (Figure. 3A). There were no other differences in Atg9a mRNA expression between healthy and arresting trophoblast tissue nor between the two gestation days studied. Atg9b mRNA expression was greater (P < 0.05) in AT than HT on gd50 (Figure 3B). As with Atg9a gene, no other differences in Atg9b expression levels were recorded in this study. These results suggest that both Atg9 genes may be involved in maintaining healthy pregnancy in swine. The elevated Atg9 gene expression levels in arresting tissue samples, as compared to the healthy trophoblast tissue, also suggest that these genes are mainly activated by unfavorable uterine conditions during pregnancy in sows; this is in agreement with observations obtained after miscarriages in women [5]. The specific reason for the differences in Atg9a mRNA and Atg9b mRNA expression between gd20 and gd50 are difficult to explain, but since alterations in only one of those genes can lead to reproductive disturbances [8], they can still be implicated in embryonic and fetal arrest in pregnant pigs. One of the reasons for this disparity could be the fact that Atg9a gene is ubiquitously expressed whereas Atg9b gene is only expressed in the placenta and the pituitary gland [12]. As of yet, no experimental studies have been conducted on the role of Atg9b gene in mammalian pregnancy.
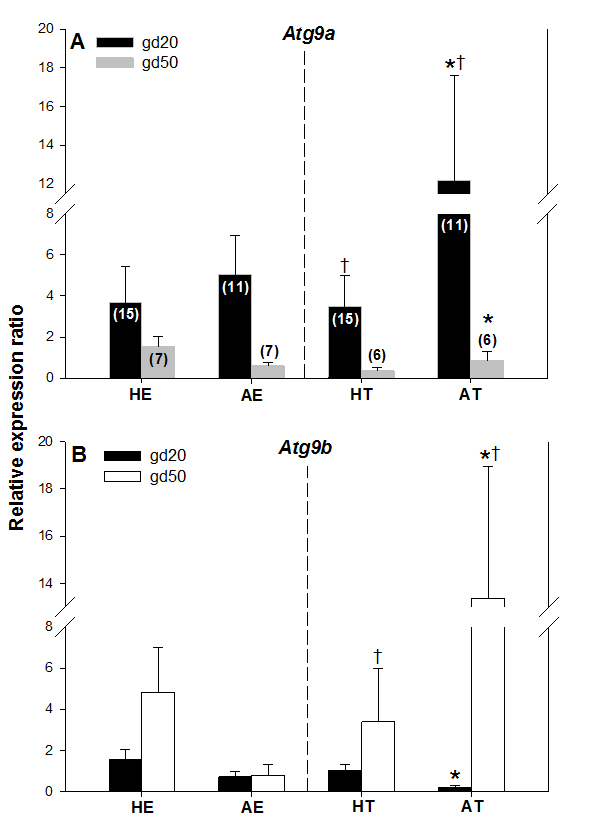
Figure 3. Comparisons of Atg9a mRNA (A) and Atg9b mRNA (B) expression levels in healthy and arresting porcine conceptuses and attachment sites on gestational days 20 (gd20) and 50 (gd50). Healthy endometrium (HE), arresting endometrium (AE), healthy trophoblast (HT), arresting trophoblast (AT). Numbers of samples used for rtPCR are given in parentheses (upper panel). Values denoted by the same symbols (*) differ significantly.
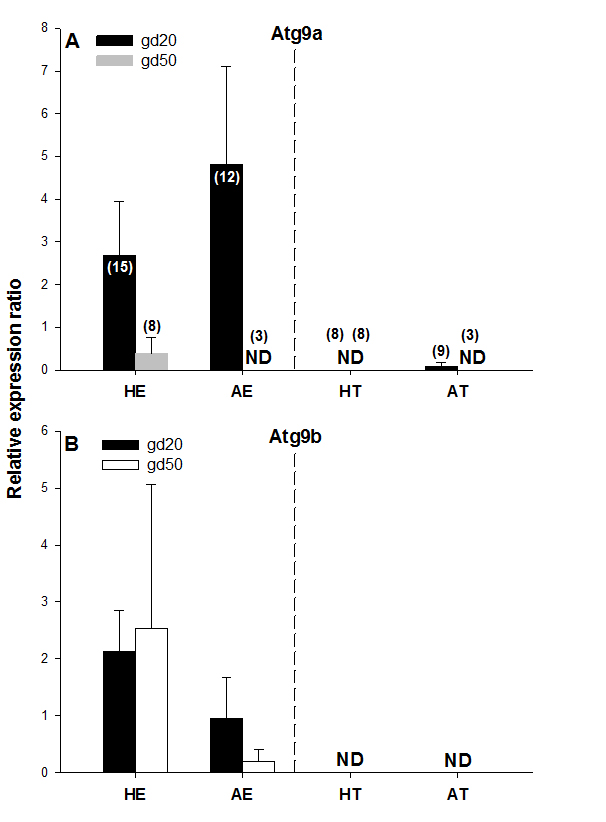
Figure 4. Atg9a and Atg9b protein expression on gestational days 20 (gd20) and 50 (gd50) in healthy and arresting conceptuses and their attachment sites. Protein expression levels were calculated as the ratios to β-actin from each corresponding blot; the bands detected in the 87-kDa range were quantified. Numbers of samples used for Western blot are given in parentheses (upper panel). Healthy endometrium (HE), arresting endometrium (AE), healthy trophoblast (HT), arresting trophoblast (AT), and non-detectable (ND).
No protein expression was detected with the Western blot of both Atg9 proteins in trophoblast tissue samples on either of the two gestational days studied except for low levels of Atg9a protein in AT samples on gd20 (Figure 4A and 4B, Figure 5). In addition, no Atg9a protein was detected in AE obtained on gd50 (Figure. 4A). On either gestational day, no variation in Atg9 protein levels between healthy or arresting conceptus attachment sites was detected. This is in contrast with previous observations in humans [13]. Autophagy was predominantly localized to the syncytiotrophoblast layer and autophagosomes were more abundant in the fetal growth restricted (FGR) placentae [13]. Moreover, the autophagy regulators including LC3B, Beclin-1, Atg5, Atg9 and Atg16L1 were all detected in villous trophoblast [14]. Our results, however, appear to be in partial agreement with a previous rodent study, wherein the absence of Atg9a gene expression resulted in fetal developmental abnormalities and demise. A lack of protein detection could be due to a number of factors including mRNA stability, protein degradation or protein relocation. Protein production depends on the abundance and stability of mRNA; rapid decays of mRNA can prevent the accumulation of transcripts and protein biosynthesis [15]. Alternatively, intrauterine proteins may have been degraded in response to various hormonal stimuli [16]. In the present Western blotting experiments, the real weight of proteins was somewhat less than the theoretical weight (positive control), which can be indicative of ongoing post-translational modifications [17]. Protein relocation could also have influenced protein detection in the present study [18, 19]. Further research is needed to provide information about synthetic/secretory pathways and specific roles of the Atg9 proteins in porcine pregnancy. It is attractive to speculate that inadequate expression of Atg proteins was associated with the altered development of conceptus in the sows of the present study. It would now be interesting to test the effect of various autophagy inhibitors (e.g., LY294002, Paclitaxel or Wortmannin) and activators (e.g., Temozolomide, Metformin or Trifluoperazine; Figure 1), administered just prior to the periods of expected embryonic or fetal arrest, on the piglet productivity.
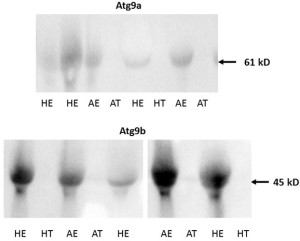
Figure 5. Examples of Western blot images containing Atg9a (upper panel) and Atg9b (lower panel) protein bands from porcine tissue specimens collected on gestational day 20. The intensity of bands for both Atg9a and Atg9b proteins were subsequently quantified relative to the expression of the beta actin bands. Healthy endometrium (HE), arresting endometrium (AE), healthy trophoblast (HT), arresting trophoblast (AT), and (+) manufacturer’s positive control.
In conclusion, there may be possible involvement of embryonic and fetal autophagy-related genes in the placentation and development of porcine conceptuses. Due mainly to the discrepancies in the results from rtPCR and Western blotting, more research is needed on the transcriptional regulation of Atg9 genes as well as the sites and mode of action of their products during porcine pregnancy.
Competing Interests: The authors declare that they have no competing interests.
Acknowledgements: Thanks are extended to: staff members at the Arkell Swine Research Station (OMAFRA) in Guelph, ON, Canada, for care and management of experimental animals; the Meat Wing of the Department of Animal and Poultry Science, U of G, for help with euthanasia; Dr. Jocelyn Wessels for technical assistance with specimen collection and the preparation of samples; Dr. Tami Martino (Department of Biomedical Sciences, U of G) for access to the rtPCR and Western blot facilities, and the Jordan University of Science and Technology (sabbatical funding for RTK). The present results were presented, in the preliminary form, during the 4th World Congress of Reproductive Biology (Okinawa, Japan; 27-29 September 2017).
Funding Information: Primary funding in the form of an operating research grant to CT was provided by the Ontario Pork. Additional funding was provided by the Department of Biomedical Sciences, U of G (PMB).
References
- Wilson ME (2002) Role of placental function in mediating conceptus growth and survival. J Anim Sci 80(E. Suppl.2) E195–E201.
- Pope WF (1994) Embryonic Mortality in Swine. In: Embryonic Mortality in Domestic Species. CRC Press 53–78.
- Glick D, Barth S, Macleod KF (2010) Autophagy: cellular and molecular mechanisms. J Pathol 221: 3–12. [crossref]
- Mizushima N, Levine B (2010) Autophagy in mammalian development and differentiation. Nat Cell Biol 12: 823–830. [crossref]
- Avagliano L, Terraneo L, Virgili E, Martinelli C, Doi P2, et al. (2015) Autophagy in Normal and Abnormal Early Human Pregnancies. Reprod Sci 22: 838–844. [crossref]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, et al. (2012) Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 198–219: 33.
- Bestebroer J, V’kovski P, Mauthe M, Reggiori F (2013) Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic 14: 1029–41.
- Kojima T, Yamada T, Akaishi R, Furuta I, Saitoh T, et al. (2015) Role of the Atg9a gene in intrauterine growth and survival of fetal mice. Reprod Biol 15: 131–138. [crossref]
- Croy BA, Wessels J, Linton N, Tayade C (2009) Comparison of immune cell recruitment and function in endometrium during development of epitheliochorial (pig) and hemochorial (mouse and human) placentas. Placenta 30(Suppl A) 2009: S26–S31.
- Bidarimath M, Khalaj K, Kridli RT, Wessels J, et al. (2017) Altered expression of chemokines and their receptors at porcine maternal-fetal interface during early and mid-gestational fetal loss. Cell Tissue Res.
- Kridli RT, Khalaj K, Bidarimath M, Tayade C (2015) Placentation, maternal-fetal interface, and conceptus loss in swine. Theriogenology 85,135–44.
- Xie Y, Kang R, Sun X, Zhong M, et al. (2015) Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 28–45.
- Bagola K, Menhert M, Jarosch E, Sommer T. Protein dislocation from the ER. Biochim Biophys Acta 2011: 1808: 25–36.
- Curtis S, Jones CJP, Garrod A, Hulme CH, et al. (2013) Identification of autophagic vacuoles and regulators of autophagy in villous trophoblast from normal term pregnancies and in fetal growth restriction. J Matern Fetal Neo M 26,339–46.
- Yamada T, Carson A, Caniggia I, Umebayashi K, et al. (2005) Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an Autophagy-related Protein (APG9-like2) highly expressed in trophoblast. J Biol Chem 18283–90.
- Wu X, Brewer G (2012) The regulation of mRNA stability in mammalian cells: 2.0. Gene 500: 10–21. [crossref]
- Cooper GM (2000) Protein Degradation. The Cell: A Molecular Approach, 2nd Edition. Sunderland: Sinauer Associates.
- Mahmood T, Yang PC (2012) Western blot: technique, theory, and trouble shooting. N Am J Med Sci 4: 429–434. [crossref]
- Bagola K, Mehnert M, Jarosch E, Sommer T (2011) Protein dislocation from the ER. Biochim Biophys Acta 1808: 925–936. [crossref]